 Author
Author  Correspondence author
Correspondence author
Triticeae Genomics and Genetics, 2024, Vol. 15, No. 2 doi: 10.5376/tgg.2024.15.0008
Received: 06 Feb., 2024 Accepted: 10 Mar., 2024 Published: 20 Mar., 2024
Zhou J., and Liu X.M., 2024, Triticeae and gluten: genetic bases of wheat allergies and sensitivities Triticeae, Triticeae Genomics and Genetics, 15(2): 77-87 (doi: 10.5376/tgg.2024.15.0008)
The Triticeae tribe, including major cereals such as wheat, oats, and rye, plays a crucial role in global agriculture and human nutrition. Gluten, the main protein complex in wheat, is indispensable in food processing due to its unique viscoelastic properties but is also a major concern for health issues such as celiac disease (CD), non-celiac wheat sensitivity (NCWS), and wheat allergies. This study aims to review the genetic bases of wheat allergies and sensitivities, including the structure and function of gluten proteins, the genetic variability of allergenic gluten proteins, and diagnostic methods. By exploring the mechanisms of NCWS and the genetic predisposition to CD, this study will also introduce the latest advances in genomic research and molecular breeding techniques, particularly the application of CRISPR/Cas9 gene editing technology in reducing wheat allergens. The goal is to integrate genomic approaches and advanced breeding techniques to develop hypoallergenic wheat varieties, thereby improving the safety and quality of wheat products to meet the growing consumer demand for allergen-free foods. This study hopes to provide new insights into the understanding of wheat allergies and sensitivities and offer scientific basis and practical guidance for developing safer and higher-quality wheat products in the future.
1 Introducion
The Triticeae tribe, including major cereal crops such as wheat (Triticum aestivum), barley (Hordeum vulgare), and rye (Secale cereale), plays a crucial role in global agriculture and human nutrition. Wheat, in particular, is a staple food for a significant portion of the world's population, providing essential nutrients and energy (Mochida and Shinozaki, 2013). The primary protein complex in wheat, gluten, is composed of gliadins and glutenins, which are responsible for the unique viscoelastic properties of dough (Zhou et al., 2021). However, gluten is also the main culprit behind several wheat-related disorders, including celiac disease (CD), non-celiac wheat sensitivity (NCWS), and wheat-dependent exercise-induced anaphylaxis (WDEIA) (Mameri et al., 2012; Ozuna and Barro, 2018).
The prevalence of gluten-related disorders has been on the rise, making it imperative to understand the genetic and immunological bases of these conditions (Ozuna and Barro, 2018). Celiac disease, an autoimmune disorder triggered by gluten ingestion, affects approximately 1% of the global population. NCWS and WDEIA, although less understood, also pose significant health risks (Altenbach et al., 2015; Lombardo et al., 2015). The complexity of these conditions is highlighted by the diverse immunological responses to different gluten proteins, such as α-gliadins, ω-gliadins, and high-molecular-weight glutenin subunits (HMW-GSs) (Altenbach et al., 2015; Cao et al., 2022). Understanding the genetic factors that influence gluten protein composition and immunogenicity can lead to the development of hypoallergenic wheat varieties and improve the quality of life for affected individuals (Rogers et al., 2004; Lombardo et al., 2015).
This study aims to comprehensively explore the genetic bases of wheat allergies and sensitivities, focusing on the Triticeae tribe. It will elucidate the genetic diversity and evolution of gluten proteins within the Triticeae tribe, analyze the immunogenic properties of different gluten proteins and their roles in wheat-related disorders, and explore recent advancements in genetic and genomic tools that facilitate the breeding of hypoallergenic wheat varieties. Additionally, the study will discuss potential strategies for mitigating the impact of gluten-related disorders through genetic modification and selective breeding. By addressing these objectives, this research seeks to provide scientific foundations for developing safer and more nutritious wheat varieties, ultimately benefiting individuals with gluten-related disorders and the broader agricultural community.
2 Gluten: Structure and Function
2.1 Composition of gluten proteins
Gluten is a complex mixture of storage proteins found in wheat and related grains. It primarily consists of two main protein groups: glutenins and gliadins. Glutenins are further divided into high molecular weight glutenin subunits (HMW-GS) and low molecular weight glutenin subunits (LMW-GS). The HMW-GS, such as 1Ax2.1* and 1By19* identified in spelt wheat, play a crucial role in the formation of gluten macropolymers due to their ability to form intermolecular disulfide bonds with other glutenins and gliadins (Cao et al., 2022). Similarly, the LMW-GS, such as the Glu-B3h gene, contribute to the overall quality of wheat by enhancing dough strength and breadmaking properties (Wang et al., 2016).
2.2 Functional properties of gluten in food
The functional properties of gluten are largely attributed to its unique viscoelastic characteristics, which are essential for the texture and structure of various wheat-based products. The HMW-GS, with their longer repetitive regions and high percentage of α-helices, are particularly beneficial for forming superior gluten macropolymers, which enhance the elasticity and extensibility of dough (Cao et al., 2022). On the other hand, the LMW-GS, such as those encoded by the Glu-B3h gene, contribute to superior flour quality by increasing dough strength and loaf volume, which are critical parameters in breadmaking (Wang et al., 2016).
2.3 The role of gluten in wheat quality
The quality of wheat is largely influenced by its gluten composition and functionality. Wang et al. (2016) used the wheat cultivar CB037B and its Glu-B3 deletion line CB037C to comprehensively analyze the molecular characteristics and functional properties of the Glu-B3h gene through proteomics and molecular biology methods. The study results showed that the absence of Glu-B3h did not significantly affect plant morphology or yield traits but led to a reduction in the number and size of protein bodies, as well as significant decreases in key quality parameters such as dough mixing performance, strength, bread volume, and scores (Figure 1). The Glu-B3h gene, identified as a low-molecular-weight glutenin subunit (LMW-GS), has been confirmed as a crucial determinant of dough strength and baking quality, further highlighting the importance of gluten composition in determining wheat quality (Wang et al., 2016). The presence of specific high-molecular-weight glutenin subunits (HMW-GS), such as 1Ax2.1 and 1By19, has been shown to improve wheat quality by forming a stronger gluten network, which is essential for producing high-quality bread and other wheat-based products (Cao et al., 2022).
 Figure 1 The comparison of GMP contents and pan bread between CB037B and CB037C (Adopted from Wang et al., 2016) Image caption: (a) GMP content determination of CB037B and CB037C at 5, 8, 11, 14, 17, 20, 23, 26, 29 DPA; (b) Separation and identification of GMP in the CB037B and CB037C by SE-HPLC; (c) Pan bread appearance of CB037B and CB037C (Adopted from Wang et al., 2016) |
Figure 1 by Wang et al. (2016) illustrates the impact of the Glu-B3h gene on gluten content and dough performance. Figures 1a and 1b show the variations in gluten content during different developmental stages in CB037B and CB037C. The study indicates that the absence of Glu-B3h significantly reduces the gluten macropolymer (GMP) content, which directly affects dough mixing performance. Figure 1c demonstrates that the lack of Glu-B3h significantly lowers the dough tolerance index, development time, stability, maximum resistance, and extensibility. Figure 1d depicts changes in bread volume and scores, showing that the absence of Glu-B3h leads to a significant decrease in both bread volume and scores. These results clearly indicate that the Glu-B3h gene, by influencing the formation of gluten macropolymers, significantly enhances dough performance and bread-making quality. The study provides direct visual evidence of the importance of the Glu-B3h gene in wheat quality improvement. It emphasizes the potential of genetic engineering and marker-assisted selection to improve gluten quality, offering new tools and methods for wheat breeding.
3 Genetic Bases of Wheat Allergies
3.1 Overview of wheat allergies
Wheat allergies are a significant health concern, affecting a substantial portion of the population. These allergies can manifest in various forms, including gastrointestinal and systemic allergic reactions, upon the ingestion of wheat. The immunological reactivity to wheat proteins, particularly gluten, is a primary factor in the etiology of these allergic responses. Recent studies have shown an increase in the frequency of gluten-related disorders, such as celiac disease (CD) and non-celiac wheat sensitivity (NCWS) (Ozuna and Barro, 2018).
3.2 Allergenic gluten proteins
Gluten proteins, which include gliadins and glutenins, are the main allergenic components in wheat. These proteins are crucial for the physico-chemical properties of bread dough and contribute significantly to the protein intake in the human diet. However, in certain individuals, these proteins trigger immunological reactions. A novel major wheat food allergen, Tri a 36, has been identified as a low molecular weight glutenin. This allergen is highly reactive with IgE antibodies from wheat food-allergic patients and shows cross-reactivity with related allergens in other cereals such as rye, barley, oat, spelt, and rice (Baar et al., 2012).
3.3 Genetic variability in allergenic proteins
The genetic variability in allergenic proteins among different wheat species and their ancestors has been a subject of extensive research. Studies have shown that domestication and breeding have led to a decrease in the content of gliadins and total gluten in cultivated Triticeae species compared to their wild ancestors. This reduction in gliadins, which are rich in immunogenic epitopes, suggests a potential decrease in allergenicity. However, the levels of glutenin subunits, including those related to Tri a 36, tend to be higher in cultivated species. The variability in the number of CD immunogenic epitopes per species and genome is significant, with higher frequencies associated with certain genomes such as DD, BBAADD, and RR types (Ozuna and Barro, 2018).
3.4 Diagnostic methods for wheat allergies
The diagnosis of wheat allergies involves both molecular and immunological approaches. The identification and characterization of specific allergens, such as Tri a 36, enable the development of precise diagnostic tools. Recombinant allergens can be used for molecular diagnosis and for designing specific immunotherapy strategies. Tri a 36, for instance, has been shown to induce specific and dose-dependent basophil activation and remains reactive even after extensive digestion, making it a reliable marker for wheat food allergy diagnosis (Baar et al., 2012).
4 Genetic Bases of Wheat Sensitivities
4.1. Overview of wheat sensitivities (Non-Celiac)
Wheat sensitivities, distinct from celiac disease, encompass a range of adverse reactions to wheat ingestion, including wheat allergy and non-celiac wheat sensitivity (NCWS). These conditions are characterized by various symptoms such as gastrointestinal distress, respiratory issues, and skin reactions, which are not mediated by the autoimmune mechanisms seen in celiac disease.
Wheat allergy is an immunological response to wheat proteins, often involving IgE antibodies. Genetic factors play a significant role in the predisposition to wheat allergies. For instance, polymorphisms in the interleukin-18 (IL-18) gene have been associated with sensitization to wheat flour in bakery workers, suggesting a genetic basis for the immune response to wheat allergens (Kim et al., 2012).
Non-celiac wheat sensitivity (NCWS) is a more ambiguous condition, lacking specific biomarkers and clear immunological pathways. However, genetic predispositions may still influence susceptibility. For instance, a study by Corsi et al. (2020) using association mapping and multi-parent advanced generation inter-cross (MAGIC) population analysis found that sensitivity to ToxB is primarily controlled by the Tsc2 locus located on chromosome 2B. The Tsc2 locus on chromosome 2B is associated with sensitivity to the ToxB effector from the pathogen Pyrenophora tritici-repentis, which causes tan spot in wheat (Figure 2). This locus and other similar loci may provide insights into the genetic basis of NCWS.
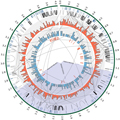 Figure 2 Circos plot illustrating key genomic and genic features of the ‘Synthetic W7984’ ‘super-contig’ (705 assembly scaffolds, totalling 3.47 Mb) spanning the wider Tsc2 locus, based on the ‘Chinese Spring’ region 2B: 14 040 000~30 500 000 bp (Adopted from Corsi et al., 2021) Image caption: Tracks, from outside to inside: (1) the super-contig, with size indicated in Mb; (2) tick marks to illustrate start/end points between scaffolds, (3) loops to indicate sequence homology of genes based on 91-mers, (4) histogram of transposable element density, (5) histogram of gene density, (6) candidate genes, (7) tick marks indicating the named SNPs that delineate the boundaries of the most likely Tsc2 region (Adopted from Corsi et al., 2021) |
Figure 2 from Corsi et al. (2020) shows a Circos plot of the synthetic "W9784" ultralong sequence within the Chinese Spring wheat region (2B: 14,040,000-30,500,000 bp). The figure presents key genomic and gene features of this region through multilayer circular plots, including 705 assembled scaffolds with a total length of 3.47 Mb, spanning the broader Tsc2 locus. The plot details sequence homology, transposon element density, gene density, and the locations of candidate genes, marked with multiple SNP markers. The circular structure of the plot visually displays the distribution and association of these genes and markers on the chromosome, providing a clear visual tool for understanding the genetic structure of the ToxB sensitivity region.
Furthermore, the gene Snn3-D1, identified in Aegilops tauschii, the D-genome progenitor of common wheat, has been implicated in susceptibility to Septoria nodorum blotch (SNB) through its interaction with the necrotrophic effector SnTox3 (Zhang et al., 2021). This gene's role in disease susceptibility highlights the complex genetic interactions that can influence wheat sensitivities.
4.2 Mechanisms of Non-Celiac wheat sensitivities
Non-celiac wheat sensitivity (NCWS) is a condition characterized by gastrointestinal and extra-intestinal symptoms related to the ingestion of wheat, in the absence of celiac disease or wheat allergy. The pathophysiological mechanisms underlying NCWS are not well understood, and the diagnosis is primarily based on the exclusion of other wheat-related disorders and the presence of symptoms that improve on a gluten-free diet.
Recent studies have begun to elucidate the genetic and molecular bases of NCWS. A genome-wide transcriptomic analysis of the intestinal mucosa from NCWS patients revealed significant differences in gene expression profiles compared to healthy controls. Specifically, 300 RNA transcripts were differentially expressed, with only 37% being protein-coding RNA and the remainder being non-coding RNA. This suggests a complex regulatory network involving non-coding RNA in the pathogenesis of NCWS. Principal component analysis and receiver operating characteristic curves indicated that these gene expression profiles could potentially distinguish NCWS patients from controls, highlighting the potential for developing diagnostic biomarkers (Efthymakis et al., 2020).
Further analysis of these differentially expressed genes pointed to the involvement of the innate immune response, the hedgehog signaling pathway, and circadian rhythm dysregulation in NCWS. These findings suggest that NCWS may result from a deranged immune response, possibly triggered by gluten or other wheat components. The role of non-coding RNA in regulating these pathways underscores the importance of regulatory genetic circuits in the disease mechanism (Efthymakis et al., 2020).
4.3 Genetic factors influencing wheat sensitivities
Wheat sensitivities, including celiac disease (CD) and non-celiac wheat sensitivity (NCWS), are influenced by various genetic factors. The immunological reactivity to gluten proteins, particularly gliadins and glutenins, plays a crucial role in these conditions. Research has shown that domestication and breeding of wheat have led to a decrease in the content of gliadins, which are known to contain immunogenic epitopes associated with CD (Ozuna and Barro, 2018). Specifically, the genomes of certain wheat species, such as those with DD, BBAADD, and RR types, have been found to have a higher frequency of CD immunogenic epitopes (Ozuna and Barro, 2018).
Moreover, the identification of genetic mutations that lower gluten content in wheat has been a significant advancement. For instance, the inactivation of a transcription factor in barley, which was then applied to wheat, resulted in a substantial reduction in gliadin and low-molecular-weight glutenin accumulation by 50% to 60% (Moehs et al., 2019). This genetic modification also increased the levels of lysine, an essential amino acid, by 33%, potentially offering a wheat alternative for individuals with gluten sensitivities (Moehs et al., 2019).
4.4 Diagnostic methods for wheat sensitivities
The diagnosis of wheat sensitivities involves both molecular and immunological approaches. One of the key diagnostic methods is the identification of specific wheat allergens that trigger immune responses in sensitive individuals. For example, Tri a 36, a low molecular weight glutenin, has been characterized as a major wheat food allergen. This allergen reacts with IgE antibodies in approximately 80% of wheat food-allergic patients and shows cross-reactivity with related allergens in other cereals such as rye, barley, and oats (Baar et al., 2012).. The molecular characterization of Tri a 36, including its resistance to digestion and heat, makes it a valuable target for diagnostic tests and the development of specific immunotherapy strategies (Baar et al., 2012).
Additionally, the quantification of CD immunogenic epitopes in gliadin sequences using techniques such as reversed-phase high-performance liquid chromatography (RP-HPLC) and database searches has provided insights into the variability of these epitopes among different wheat species (Ozuna and Barro, 2018). This variability is crucial for understanding the genetic predisposition to wheat sensitivities and for developing diagnostic tools that can accurately identify individuals at risk.
5 Celiac Disease and Wheat Genetics
5.1 Pathophysiology of celiac disease
Celiac disease (CD) is a chronic autoimmune disorder triggered by the ingestion of gluten, a group of proteins found in wheat, barley, and rye. The disease primarily affects the small intestine, leading to inflammation and villous atrophy, which impairs nutrient absorption. Gluten proteins, particularly gliadins and glutenins in wheat, are resistant to gastrointestinal digestion, resulting in the formation of immunogenic peptides. These peptides are presented by antigen-presenting cells to T cells, leading to an inflammatory response that damages the intestinal mucosa (Moehs et al., 2019; Picascia et al., 2020).
5.2 Genetic predisposition to celiac disease
The genetic predisposition to celiac disease is strongly associated with specific human leukocyte antigen (HLA) class II genes, particularly HLA-DQ2 and HLA-DQ8. Approximately 95% of individuals with celiac disease carry the HLA-DQ2 haplotype, while most of the remaining patients carry HLA-DQ8. These HLA molecules present gluten-derived peptides to CD4+ T cells, initiating the autoimmune response. However, the presence of these HLA alleles alone is not sufficient to cause the disease, indicating that other genetic and environmental factors also play a role in its pathogenesis (Moehs et al., 2019; Picascia et al., 2020).
5.3 Gluten proteins and celiac disease
Gluten proteins, including gliadins and glutenins in wheat, hordeins in barley, and secalins in rye, are rich in proline and glutamine residues, making them resistant to complete digestion by gastrointestinal enzymes. The resulting peptides can cross the intestinal epithelium and be deamidated by tissue transglutaminase, enhancing their binding affinity to HLA-DQ2 and HLA-DQ8 molecules. This process is crucial for the activation of gluten-specific T cells and the subsequent inflammatory response. Recent research has focused on developing wheat varieties with reduced immunogenic gluten sequences to mitigate the adverse effects in celiac patients (Moehs et al., 2019; Picascia et al., 2020).
5.4 Diagnostic methods for celiac disease
The diagnosis of celiac disease typically involves a combination of serological tests and intestinal biopsy. Serological tests detect specific antibodies, such as anti-tissue transglutaminase (tTG) and anti-endomysial antibodies (EMA), which are highly sensitive and specific for celiac disease. Positive serological tests are usually followed by an intestinal biopsy to confirm the diagnosis, which reveals characteristic histological changes such as villous atrophy, crypt hyperplasia, and increased intraepithelial lymphocytes. Additionally, genetic testing for HLA-DQ2 and HLA-DQ8 can support the diagnosis, especially in cases with inconclusive serology or biopsy results (Moehs et al., 2019; Picascia et al., 2020).
6 Advances in Genetic Research on Wheat Allergies and Sensitivities
6.1 Genomic approaches to understanding wheat allergies
Recent advancements in genomic technologies have significantly enhanced our understanding of wheat allergies and sensitivities. The advent of next-generation sequencing (NGS) has allowed for the comprehensive mapping of the wheat genome, facilitating the identification of genes associated with allergenic responses. For instance, the International Wheat Genome Sequencing Consortium has provided a fully annotated reference wheat-genome assembly, which has been instrumental in identifying and characterizing allergenic proteins such as prolamins and non-prolamin allergens (Juhász et al., 2018; Hussain et al., 2022). These genomic resources have enabled researchers to pinpoint specific genetic variations and expression patterns linked to wheat allergies, thereby offering new avenues for developing hypoallergenic wheat varieties.
6.2 Molecular breeding for reduced allergenicity
Molecular breeding techniques, including marker-assisted selection (MAS) and genomic selection (GS), have been employed to develop wheat varieties with reduced allergenicity. By leveraging the extensive genomic data available, breeders can now select for traits that minimize the presence of allergenic proteins. For example, RNA interference (RNAi) has been used to silence the γ-gliadin genes, which are major sensitizing allergens in wheat-dependent exercise-induced anaphylaxis (WDEIA). This approach has shown promise in reducing the immunogenicity of wheat, although the complexity of immune responses among individuals suggests that further refinement is needed (Altenbach et al., 2015). Additionally, the integration of omics technologies, such as proteomics and metabolomics, has provided deeper insights into the molecular mechanisms underlying wheat allergenicity, thereby enhancing the effectiveness of molecular breeding strategies (Alotaibi et al., 2020).
6.3 Genetic engineering and CRISPR/Cas9 technology
The CRISPR/Cas9 genome editing technology has revolutionized the field of genetic engineering, providing precise and efficient tools to modify the wheat genome to reduce allergenicity. CRISPR/Cas9 has been successfully used to target and edit specific genes associated with allergenic proteins. For example, combining CRISPR/Cas9 with microspore technology can rapidly generate homozygous mutant plants, thereby accelerating the breeding process (Bhowmik et al., 2018). Bhowmik et al. (2018) demonstrated the ability of CRISPR/Cas9 technology to introduce both exogenous and endogenous gene mutations in wheat microspores, providing strong evidence of how genetic engineering and CRISPR/Cas9 technology can accelerate the breeding process (Figure 3). Advances in CRISPR technology, such as base editing and prime editing, have further increased the precision and scope of gene modification, paving the way for the development of low-allergenicity wheat varieties (Li et al., 2021; Jogam et al., 2021).
 Figure 3 CRISPR/Cas9-based editing of an endogenous ubiquitin gene in wheat microspores (Adopted from Bhowmik et al., 2018) Image caption: (A) Schematic diagram of the pEEE005-NLSCas9/gUbi1 and pEEE006-NLSCas9/gUbi1/GFP constructs used for mutagenesis of in wheat microspores; (B) PCR/restriction enzyme assay to detect mutations in TaUbiL1 induced by Cas9/ gUbi1; (C) Deletions detected at the cleavage site in sequenced products of all three homoeologs of TaUbiL1 from wheat microspores transfected with pEEE005-NLSCas9/gUbi1 or pEEE006-NLSCas9/gUbi1/GFP constructs; (D) Sanger sequencing electropherograms showing mutations in TaUbiL1 homoeologues from the A, B and D genomes of wheat (Adopted from Bhowmik et al., 2018) |
Figure 3 by Bhowmik et al. (2018) shows the results of editing the endogenous ubiquitin gene (TaUbiL1) in wheat microspores using the CRISPR/Cas9 system. These images and charts display various insertion and deletion mutations, demonstrating the introduction of mutations through non-homologous end joining (NHEJ) repair following double-strand breaks. This result further proves the high efficiency of the CRISPR/Cas9 system in wheat gene editing, particularly in the application of endogenous gene editing. Utilizing Figure 3, the potential application of CRISPR/Cas9 technology in improving wheat traits, enhancing resistance, and increasing adaptability can be effectively demonstrated.
In summary, the integration of genomic approaches, molecular breeding, and cutting-edge genetic engineering techniques like CRISPR/Cas9 holds great promise for mitigating wheat allergies and sensitivities. These advancements not only contribute to the development of safer wheat products but also enhance our overall understanding of the genetic bases of wheat allergenicity.
7 Implications for Wheat Breeding and Food Industry
7.1 Breeding strategies for allergen-free wheat
Breeding strategies aimed at developing allergen-free wheat focus on reducing or eliminating the presence of immunogenic gluten proteins, particularly gliadins and glutenins, which are implicated in celiac disease (CD) and non-celiac wheat sensitivity (NCWS). Recent studies have shown that domestication and breeding have contributed to a decrease in the content of gliadins in cultivated Triticeae species compared to their wild ancestors (Ozuna and Barro, 2018). This reduction in gliadins, which are rich in CD immunogenic epitopes, suggests that selective breeding can be an effective strategy for developing wheat varieties with lower allergenic potential. Additionally, the identification of specific alleles in wild wheat relatives, such as Triticum urartu, which exhibit diverse and potentially beneficial traits for wheat improvement, provides a valuable genetic resource for breeding programs (Talini et al., 2019).
7.2 Impact on food production and quality
The reduction of gluten proteins in wheat through breeding strategies has significant implications for food production and quality. Gluten proteins are crucial for the viscoelastic properties of dough, which affect the texture and quality of bread and other wheat-based products. While reducing gliadins can lower the allergenic potential of wheat, it is essential to balance this with the need to maintain or improve the functional properties of wheat flour. Studies have shown that cultivated barley, which has lower immunogenic epitopes and gluten content, could serve as an alternative cereal with favorable properties for food production (Ozuna and Barro, 2018). Furthermore, the diverse allelic pool of wild wheat relatives, such as T. urartu, offers opportunities to enhance wheat quality traits, including flour quality and carotenoid concentrations, which can contribute to the development of high-quality, allergen-free wheat products (Talini et al., 2019).
7.3 Regulatory and safety considerations
The development and commercialization of allergen-free wheat varieties must adhere to stringent regulatory and safety standards to ensure consumer safety. Regulatory agencies require comprehensive testing to confirm the absence or significant reduction of immunogenic epitopes in new wheat varieties. This includes molecular characterization, immunological assays, and clinical trials to assess the safety and efficacy of the allergen-free wheat. The variability in the number of CD epitopes among different Triticeae species and genomes highlights the need for precise and thorough evaluation of each new variety (Ozuna and Barro, 2018). Additionally, regulatory frameworks must address labeling requirements to inform consumers about the allergen-free status of wheat products, ensuring transparency and trust in the marketplace.
7.4 Market potential and consumer acceptance
The market potential for allergen-free wheat is substantial, given the increasing prevalence of gluten-related disorders and the growing demand for gluten-free products. Consumers with CD, NCWS, and wheat allergies are actively seeking safe and nutritious alternatives to traditional wheat products. The development of allergen-free wheat varieties that retain desirable agronomic and quality traits can meet this demand and expand market opportunities. Consumer acceptance will depend on the sensory qualities, nutritional value, and safety of the new wheat products. Effective communication and education about the benefits and safety of allergen-free wheat will be crucial in gaining consumer trust and acceptance. The diverse genetic resources available in wild wheat relatives, such as T. urartu, provide a promising foundation for breeding programs aimed at meeting these market needs (Talini et al., 2019).
8 Conclusion Remarks
Research on the Triticeae tribe and gluten has revealed significant findings regarding the genetic bases of wheat allergies and sensitivities. Domestication and breeding have reduced the content of gliadins in cultivated Triticeae species, which are major components responsible for triggering celiac disease (CD) and non-celiac wheat sensitivity (NCWS). Genetic modifications, such as the inactivation of the lys3a gene in barley, have successfully lowered the accumulation of gliadins and low-molecular-weight glutenins in modified wheat lines, providing new avenues for developing hypoallergenic wheat. Additionally, the discovery of new allergens like Tri a 36 highlights the complexity of wheat allergies and underscores the need for improved diagnostic tools and specific immunotherapy strategies. The identification of new high-molecular-weight glutenin subunits in spelt wheat, such as 1Ax2.1 and 1By19, offers new genetic resources for improving wheat quality.
However, these studies also present several challenges and opportunities. The genetic complexity of gluten proteins and their immunogenic epitopes complicates the development of hypoallergenic wheat varieties. Genetically modified wheat may face consumer resistance, and public perception and regulatory hurdles need to be addressed. Furthermore, the identification of new allergens like Tri a 36 necessitates the development of better diagnostic tools and targeted immunotherapy strategies. The integration of new glutenin genes from spelt wheat into bread wheat breeding programs offers opportunities to enhance wheat quality but requires careful selection and validation in breeding populations.
Future research should focus on comprehensive genomic studies to identify and characterize all potential immunogenic epitopes in gluten proteins across different Triticeae species. Advanced breeding techniques, such as CRISPR/Cas9, should be utilized to precisely edit genes associated with gluten content and allergenicity. Further characterization of newly identified allergens and the exploration of integrating novel glutenin genes into commercial wheat varieties are also crucial. Through these efforts, safer and higher-quality wheat products can be developed, benefiting individuals with wheat allergies and sensitivities.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Alotaibi F., Alharbi S., Alotaibi M., Mosallam M., Motawei M., and Alrajhi A., 2020, Wheat omics: classical breeding to new breeding technologies, Saudi Journal of Biological Sciences, 28: 1433-1444.
https://doi.org/10.1016/j.sjbs.2020.11.083
Altenbach S., Tanaka C., Pineau F., Lupi R., Drouet M., Beaudouin E., Morisset M., and Denery-Papini S., 2015, Assessment of the allergenic potential of transgenic wheat, (Triticum aestivum) with reduced levels of ω5-gliadins the major sensitizing allergen in wheat-dependent exercise-induced anaphylaxis, Journal of Agricultural and Food Chemistry, 63(42): 9323-9332.
https://doi.org/10.1021/acs.jafc.5b03557
Baar A., Pahr S., Constantin C., Scheiblhofer S., Thalhamer J., Giavi S., Papadopoulos N., Ebner C., Mari A., Vrtala S., and Valenta R., 2012, Molecular and immunological characterization of Tri a 36 a low molecular weight glutenin as a novel major wheat food allergen, The Journal of Immunology, 189(6): 3018-3025.
https://doi.org/10.4049/jimmunol.1200438
Bhowmik P., Ellison E., Polley B., Bollina V., Kulkarni M., Ghanbarnia K., Song H., Gao C., Voytas D., and Kagale S., 2018, Targeted mutagenesis in wheat microspores using CRISPR/Cas9, Scientific Reports, 8(1): 6502.
https://doi.org/10.1038/s41598-018-24690-8
PMid:29695804 PMCid:PMC5916876
Cao Y., Zhang J., Wang R., Sun H., and Yan Y., 2022, Molecular Characterization and SNP-based molecular marker development of two novel high molecular weight glutenin genes from Triticum spelta L., International Journal of Molecular Sciences, 23(19): 11104.
https://doi.org/10.3390/ijms231911104
PMid:36232404 PMCid:PMC9570065
Corsi B., Percival‐Alwyn L., Downie R., Venturini L., Iagallo E., Mantello C., McCormick-Barnes C., See P., Oliver R., Moffat C., and Cockram J., 2020, Genetic analysis of wheat sensitivity to the ToxB fungal effector from Pyrenophora tritici-repentis the causal agent of tan spot, Theoretical and Applied Genetics, 133: 935-950.
https://doi.org/10.1007/s00122-019-03517-8
PMid:31915874 PMCid:PMC7021774
Efthymakis K., Clemente E., Marchioni M., Nicola M., Neri M., and Sallese M., 2020, An exploratory gene expression study of the intestinal mucosa of patients with non-celiac wheat sensitivity, International Journal of Molecular Sciences, 21(6): 1969.
https://doi.org/10.3390/ijms21061969
PMid:32183058 PMCid:PMC7139384
Hussain B., Akpınar B., Alaux M., Algharib A., Sehgal D., Ali Z., Aradottir G., Batley J., Bellec A., Bentley A., Cagirici H., Cattivelli L., Choulet F., Cockram J., Desiderio F., Devaux P., Doğramacı M., Dorado G., Dreisigacker S., Edwards D., El-Hassouni K., Eversole K., Fahima T., Figueroa M., Gálvez S., Gill K., Govta L., Gul A., Hensel G., Hernández P., Crespo-Herrera L., Ibrahim A., Kilian B., Korzun V., Krugman T., Li Y., Liu S., Mahmoud A., Morgounov A., Muslu T., Naseer F., Ordon F., Paux E., Perović D., Reddy G., Reif J., Reynolds M., Roychowdhury R., Rudd J., Sen T., Sukumaran S., Ozdemir B., Tiwari V., Ullah N., Unver T., Yazar S., Appels R., and Budak H., 2022, Capturing wheat phenotypes at the genome level, Frontiers in Plant Science, 13: 851079.
https://doi.org/10.3389/fpls.2022.851079
PMid:35860541 PMCid:PMC9289626
Jogam P., Sandhya D., Kumar P., Allini V., Abbagani S., and Alok A., 2021, Genetic transformation methods and advancement of CRISPR/Cas9 technology in wheat, Nanobiotechnology for Plant Protection, 2021: 253-275.
https://doi.org/10.1016/B978-0-12-821910-2.00017-5
Juhász A., Belova T., Florides C., Maulis C., Fischer I., Gell G., Birinyi Z., Ong J., Keeble-Gagnère G., Maharajan A., Ma W., Gibson P., Jia J., Lang D., Mayer K., Spannagl M., Tye-Din J., Appels R., and Olsen O., 2018, Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat, Science Advances, 4(8): eaar8602.
https://doi.org/10.1126/sciadv.aar8602
PMid:30128352 PMCid:PMC6097586
Kim S., Hur G., Jin H., Choi H., and Park H., 2012, Effect of interleukin-18 gene polymorphisms on sensitization to wheat flour in bakery workers, Journal of Korean Medical Science, 27(4): 382-387.
https://doi.org/10.3346/jkms.2012.27.4.382
PMid:22468101 PMCid:PMC3314850
Li J., Li Y., and Ma L., 2021, Recent advances in CRISPR/Cas9 and applications for wheat functional genomics and breeding, aBIOTECH, 2: 375-385.
https://doi.org/10.1007/s42994-021-00042-5
PMid:36304421 PMCid:PMC9590522
Lombardo C., Bolla M., Chignola R., Senna G., Rossin G., Caruso B., Tomelleri C., Cecconi D., Brandolini A., and Zoccatelli G., 2015, Study on the Immunoreactivity of Triticum monococcum, Einkorn) Wheat in patients with wheat-dependent exercise-induced anaphylaxis for the production of hypoallergenic foods., Journal of Agricultural and Food Chemistry 63(37): 8299-8306.
https://doi.org/10.1021/acs.jafc.5b02648
PMid:26332577
Mameri H., Bouchez I., Pecquet C., Raison-Peyron N., Choudat D., Chabane H., Kerre S., Denery-Papini S., Gohon Y., Briozzo P., Lauriére M., and Snégaroff J., 2012, A recombinant ω-gliadin-like D-type glutenin and an α-gliadin from wheat, Triticum aestivum: two immunoglobulin E binding proteins useful for the diagnosis of wheat-dependent allergies., Journal of Agricultural and Food Chemistry, 60(32): 8059-8068.
https://doi.org/10.1021/jf301992w
PMid:22809016
Moehs C., Austill W., Holm A., Large T., Loeffler D., Mullenberg J., Schnable P., Skinner W., Boxtel J., Wu L., and McGuire C., 2019, Development of decreased-gluten wheat enabled by determination of the genetic basis of lys3a Barley1, Plant Physiology, 179: 1692-1703.
https://doi.org/10.1104/pp.18.00771
PMid:30696748 PMCid:PMC6446766
Mochida K., and Shinozaki K., 2013, Unlocking Triticeae genomics to sustainably feed the future, Plant and Cell Physiology, 54(12): 1931-1950.
https://doi.org/10.1093/pcp/pct163
Ozuna C., and Barro F., 2018, Characterization of gluten proteins and celiac disease-related immunogenic epitopes in the Triticeae: cereal domestication and breeding contributed to decrease the content of gliadins and gluten, Molecular Breeding, 38: 1-16.
https://doi.org/10.1007/s11032-018-0779-0
Picascia S., Camarca A., Malamisura M., Mandile R., Galatola M., Cielo D., Gazza L., Mamone G., Auricchio S., Troncone R., Greco L., Auricchio R., and Gianfrani C., 2020, In celiac disease patients the in vivo challenge with the diploid Triticum monococcum elicits a reduced immune response compared to hexaploid wheat, Molecular Nutrition and Food Research, 64(11): 1901032.
https://doi.org/10.1002/mnfr.201901032
PMid:32374905
Rogers W., Miller T., Payne P., Seekings J., Sayers E., Holt L., and Law C., 2004, Introduction to bread wheat, Triticum aestivum L. and assessment for bread-making quality of alleles from T. boeoticum Boiss. ssp. thaoudar at Glu-A1 encoding two high-molecular-weight subunits of glutenin, Euphytica, 93: 19-29.
https://doi.org/10.1023/A:1002991206350
Talini R., Brandolini A., Miculan M., Brunazzi A., Vaccino P., Pè M., and Dell’Acqua M., 2019, Genome wide association study of agronomic and quality traits in a world collection of the wild wheat relative Triticum urartu., The Plant Journal : for Cell and Molecular Biology, 102(3): 555-568.
https://doi.org/10.1111/tpj.14650
PMid:31826330
Wang Y., Zhen S., Luo N., Han C., Lu X., Li X., Xia X., He Z., and Yan Y., 2016, Low molecular weight glutenin subunit gene Glu-B3h confers superior dough strength and breadmaking quality in wheat, Triticum aestivum L., Scientific Reports, 6(1): 27182.
https://doi.org/10.1038/srep27182
PMid:27273251 PMCid:PMC4895167
Zhang Z., Running K., Seneviratne S., Haugrud A., Szabó-Hevér A., Shi G., Brueggeman R., Xu S., Friesen T., and Faris J., 2021, A protein kinase-major sperm protein gene hijacked by a necrotrophic fungal pathogen triggers disease susceptibility in wheat, The Plant Journal, 106(3): 720-732.
https://doi.org/10.1111/tpj.15194
PMid:33576059
Zhou Z., Zhang Z., Jia L., Qiu H., Guan H., Liu C., Qin M., Wang Y., Li W., Yao W., Wu Z., Tian B., and Lei Z., 2021, Genetic basis of gluten aggregation properties in wheat, (Triticum aestivum L.) dissected by QTL mapping of glutopeak parameters, Frontiers in Plant Science, 11: 611605.
https://doi.org/10.3389/fpls.2020.611605
PMid:33584755 PMCid:PMC7876098
.png)
. PDF(770KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jin Zhou
. Xuemei Liu
Related articles
. Triticeae tribe
. Gluten
. Wheat allergies
. Non-celiac wheat sensitivity
. CRISPR/Cas9
Tools
. Email to a friend
. Post a comment


